Likelihood Tutorial
The detection, flux determination and spectral modeling of Fermi LAT sources is accomplished by a maximum likelihood optimization technique, as described in the Cicerone (see also e.g. Abdo, A. A. et al. 2009, ApJS, 183, 46). To illustrate how to use the Likelihood software, this narrative gives a step-by-step description for performing an unbinned likelihood analysis.
Unbinned vs Binned Likelihood
Unbinned likelihood analysis is the preferred method for time series analysis of the LAT data, where the number of events in each time bin is expected to be small (see Cicerone). However, for large time bins, analysis which includes bright background sources (such as the Galactic plane), and long time-baseline spectral and spatial analyses, a binned analysis is recommended. To perform a binned likelihood analysis see the Binned Likelihood tutorial.
Additional references:
Descriptions of available Spectral and Spatial Models
Examples of XML Model Definitions for Likelihood
Prerequisites
You will need an event data file, a spacecraft data file (also referred to as the `pointing and livetime history' file), and the current background models (available for download). You may choose to select your own data files, or to use the files provided within this tutorial. Custom data sets may be retrieved from the Lat Data Server.
Steps
Since there is computational overhead for each event associated with each diffuse component, it is useful to filter out any events that are not within the extraction region used for the analysis.
These simple FITS images let us inspect our data and help to pick out obvious candidate sources.
This is needed for analyzing diffuse sources, and derived absolute fluxes.
The recommended models are gll_iem_v05.fits.fits (a very large file) and iso_source_v05.txt. All of the background models along with a description of the models are available here.
The source model XML file contains the various sources and their model parameters to be fit using the gtlike tool.
Skip step 6 unless you are using background models that do not have precomputed responses.
Each event must have a separate response precomputed for each diffuse component in the source model. The precomputed responses for Pass 7 (V6) data are for the gll_iem_v05, iso_source_v05.txt, and iso_clean_05.txt diffuse models.
-
Fitting the data to the model provides flux, errors, spectralindices, and other information.
These are used for point source localization and for finding weaker sources after the stronger sources have been modeled.
1. Make Subselections from the event data
For this case the original extraction of data from the first six months of the mission was done as described in the Extract LAT Data tutorial.
Selection of data:
Search Center (RA, DEC) =(193.98, -5.82)
Radius = 20 degrees
Start Time (MET) = 239557417 seconds (2008-08-04 T15:43:37)
Stop Time (MET) = 255398400 seconds (2008-02-04 T00:00:00)
Minimum Energy = 100 MeV
Maximum Energy = 100000 MeV
We provide the user with the original photon and spacecraft data files extracted in the Extract LAT data tutorial:
If you need to combine multiple events files for your analysis, you must first generate a text file listing the events files to be included. prompt> ls *_PH* > events.txt
This text file (events.txt) will be used in place of the input fits filename when running gtselect. The syntax requires that you use an @ before the filename to indicate that this is a text file input rather than a fits file.
When analyzing a point source, it is recommended that you include events with high probability of being photons. To do this, you should use gtselect to cut on the event class, keeping only the Source class events (event class 2) (or as recommended in the Cicerone):
prompt> gtselect evclass=2
Be aware that evclass is a hidden parameter. So to use it for a cut, you must type it in the command line.
We perform a selection to filter the data we want to analyze. For this example, we consider the diffuse class photons within a 20 degree region of interest (ROI) centered on the blazar 3C 279. We apply the gtselect tool to the data file as follows:
prompt> gtselect evclass=2
Input FT1 file[] @events.txt
Output FT1 file[] 3C279_region_filtered.fits
RA for new search center (degrees) (0:360) [] 193.98
Dec for new search center (degrees) (-90:90) [] -5.82
radius of new search region (degrees) (0:180) [] 20
start time (MET in s) (0:) [] 239557417
end time (MET in s) (0:) [] 255398400
lower energy limit (MeV) (0:) [] 100
upper energy limit (MeV) (0:) [] 100000
maximum zenith angle value (degrees) (0:180) [] 100
Done.
prompt> In the last step we also selected the energy range and the maximum zenith angle value (100 degrees) as suggested in Cicerone. The Earth's limb is a strong source of background gamma rays. We filter them out with a zenith-angle cut. The value of 100 degrees is the one recommended by the LAT instrument team. The filtered data are provided here.
After the data selection is made you must compensate the livetime for the fact that you have removed some events. There are several options for calculating livetime depending on your observation type and science goals. For a detailed discussion of these options, see Likelihood Livetime and Exposure in the Cicerone. To deal with the cut on zenith-angle in this analysis, we use the gtmktime tool to exclude the time intervals where the zenith cut intersects the ROI from the list of good time intervals (GTIs). This is especially important if you are studying a narrow ROI (with a radius of less than 20 degrees), as your source of interest may come quite close to the Earth's limb. To make this correction you have to run gtmktime and answer `yes' at:
Apply ROI-based zenith angle cut[] yes
gtmktime also provides an opportunity to select GTIs by filtering on information provided in the spacecraft file. The current gtmktime filter expression recommended by the LAT team in the Cicerone is
(DATA_QUAL>0)&&(LAT_CONFIG==1)
This excludes time periods when some spacecraft event has affected the quality of the data, ensures the LAT instrument was in normal science data-taking mode, and requires that the spacecraft be within the range of rocking angles used during nominal sky-survey observations.
Here is an example of running gtmktime for our analysis of the region surrounding 3C 279.
prompt> gtmktime
Spacecraft data file[] spacecraft.fits
Filter expression[] (DATA_QUAL>0)&&(LAT_CONFIG==1)
Apply ROI-based zenith angle cut[] yes
Event data file[] 3C279_region_filtered.fits
Output event file name[] 3C279_region_filtered_gti.fitsThe data with all the cuts described above is provided in this link. A more detailed discussion of data selection can be found in the Data Preparation analysis thread.
To view the DSS keywords in a given extension of a data file, use the gtvcut tool and review the data cuts on the EVENTS extension. This provides a listing of the keywords reflecting each cut applied to the data file and their values, including the entire list of GTIs. Here we use the option `suppress_gtis=yes' to keep the output easily readable:
prompt> gtvcut suppress_gtis=yes
Input FITS file[] 3C279_region_filtered_gti.fits
Extension name[EVENTS]
DSTYP1: POS(RA,DEC)
DSUNI1: deg
DSVAL1: CIRCLE(193.98,-5.82,20)
DSTYP2: TIME
DSUNI2: s
DSVAL2: TABLE
DSREF2: :GTI
GTIs: (suppressed)
DSTYP3: ENERGY
DSUNI3: MeV
DSVAL3: 100:100000
DSTYP5: ZENITH_ANGLE
DSUNI5: deg
DSVAL5: 0:100
prompt> Here you can see the location and radius of the data selection, as well as the energy range in MeV, the zenith angle cut, and the fact that the time cuts to be used in the exposure calculation are defined by the GTI table.
Various science tools will be unable to run if you have multiple copies of a particular DSS keyword. This can happen if the position used in extracting the data from the data server is different than the position used with gtselect. It is wise to review the keywords for duplicates before proceeding. If you do have keyword duplication, it is advisable to regenerate the data file with consistent cuts.
2. Make a counts map from the event data
Next, we create a counts map of the ROI, summed over photon energies, in order to identify candidate sources and to ensure that the field looks sensible as a simple sanity check. For creating the counts map, we will use the gtbin tool with the option `CMAP' as shown below:
prompt> gtbin
This is gtbin version ScienceTools-v9r23p1-fssc-20110730
Type of output file
(CCUBE|CMAP|LC|PHA1|PHA2) [] CMAP
Event data file name[] 3C279_region_filtered_gti.fits
Output file name[] 3C279_region_filtered_gti_cmap.fits
Spacecraft data file name[NONE]
Size of the X axis in pixels[] 160
Size of the Y axis in pixels[] 160
Image scale (in degrees/pixel)[] 0.25
Coordinate system (CEL - celestial, GAL -galactic) (CEL|GAL)[] CEL
First coordinate of image center in degrees (RA or galactic l)[] 193.98
Second coordinate of image center in degrees (DEC or galactic b)[]-5.82
Rotation angle of image axis, in degrees[] 0.0
Projection method
e.g. AIT|ARC|CAR|GLS|MER|NCP|SIN|STG|TAN:[] AIT
prompt> ds9 3C279_region_filtered_gti_cmap.fitsThe last command launches the visualization tool ds9 and produces this display:
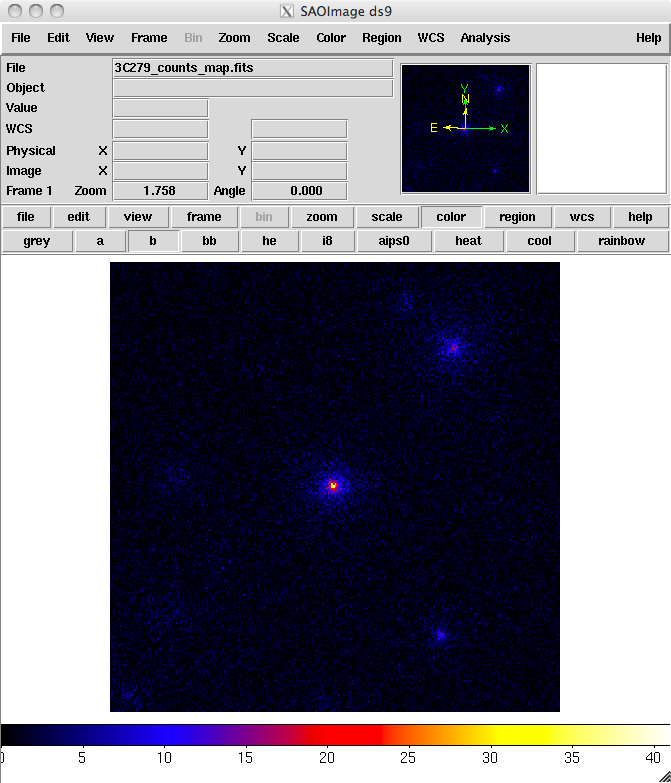
3C279_region_filtered_gti_cmap.fits
You can see one strong source and several lesser sources in this map. Mousing over the positions of these sources shows that two of them are likely 3C 279 and 3C 273. It is important to inspect your data prior to proceeding to verify that the contents are as you expect. A malformed data query or improper data selection can generate a non-circular region, or a file with zero events. By inspecting your data prior to analysis, you have an opportunity to detect such issues early in the analysis. (A more detailed discussion of data exploration can be found in the Explore LAT Data analysis thread.)
3. Make an exposure map
We are now ready to create an exposure map. The type of exposure map used by Likelihood differs significantly from the usual notion of exposure maps, which areessentially integrals of effective area over time. The exposure calculation that Likelihood uses consists of an integral of the total response over the entire ROI. See more information in the Cicerone.
Since the exposure calculation involves an integral over the ROI, separate exposure maps must be made for every distinct set of DSS cuts. This is important if, for example, one wants to subdivide an observation to look for secular flux variations from a particular source or sources. To view the DSS keywords in a given extension of a data file, use the gtvcut tool and review the data cuts for the EVENTS extension.
a. Generate a livetime cube
There are two tools needed for generating exposure maps. The first is gtltcube. This tool creates a livetimecube, which is a HealPix table, covering the full sky, of the integrated livetime as a function of inclination with respect to the LAT z-axis. For more information about the livetime cubes see the documentation in Cicerone.
Here is the example of how to run gtltcube (this step takes several minutes):
prompt> gtltcube
Event data file[] 3C279_region_filtered_gti.fits
Spacecraft data file[] spacecraft.fits
Output file[] 3C279_ltcube.fits
Step size in cos(theta) (0.:1.) [] 0.025
Pixel size (degrees)[] 1
Working on file spacecraft.fits
.....................!
prompt> Note: Values such as 0.1 for `Step size in cos(theta)' are known to give unexpected results. Use '0.09' instead.
The livetime cube generated for this analysis can be found here.
Since gtltcube produces a FITS file covering the entire sky, the output of this tool could be used for generating exposure maps for ROIs in other parts of the sky that have the same time interval selections. But use caution! The livetime cube MUST be regenerated if you change any part of the time interval selection. This can occur by changing the start or stop time of the events, or simply by changing the ROI selection or zenith angle cut (as these produce an ROI-dependent set of GTIs from gtmktime). See e.g. Data preparation in the Cicerone or the Data Preparation analysis thread.
Although the gtexpmap application (see below) can generate exposure maps for Likelihood without a livetime file, using one affords a substantial time savings.
b. Generate an exposure map.
The tool gtexpmap creates an exposure map based on the event selection used on the input photon file and the livetime cube. The exposure map must be recalculated if the ROI, zenith, energy selection or the time interval selection of the events is changed. For more information about the exposure maps see the documentation in the Cicerone.
Creating the exposure map using the gtexpmap tool, we have (this step can also take several minutes):
prompt> gtexpmap
The exposure maps generated by this tool are meant
to be used for *unbinned* likelihood analysis only.
Do not use them for binned analyses.
Event data file[] 3C279_region_filtered_gti.fits
Spacecraft data file[] spacecraft.fits
Exposure hypercube file[] 3C279_ltcube.fits
output file name[] 3C279_expmap.fits
Response functions[] CALDB
Radius of the source region (in degrees)[] 30
Number of longitude points (2:1000) [] 120
Number of latitude points (2:1000) [] 120
Number of energies (2:100) [] 20
Computing the ExposureMap using 3C279_ltcube.fits
....................!
prompt> Note that we have chosen a 30 degree radius 'source region', while the acceptance cone radius specified for gtselect was 20 degrees. See the discussion of region selection in the Cicerone. This is necessary to ensure that events from sources outside the ROI are accounted for owing to the size of the instrument point-spread function (PSF). Half-degree pixels are a nominal choice; smaller pixels should result in a more accurate evaluation of the diffuse source fluxes but will also make the exposure map calculation itself lengthier. The number of energies specifies the number of logarithmically spaced intervals bounded by the energy range given in the DSS keywords. A general recommendation is 10 bins per decade. This is sufficient to accommodate the change in effective area with energy near 100 MeV.
Here is one image plane of the exposure map we just created:
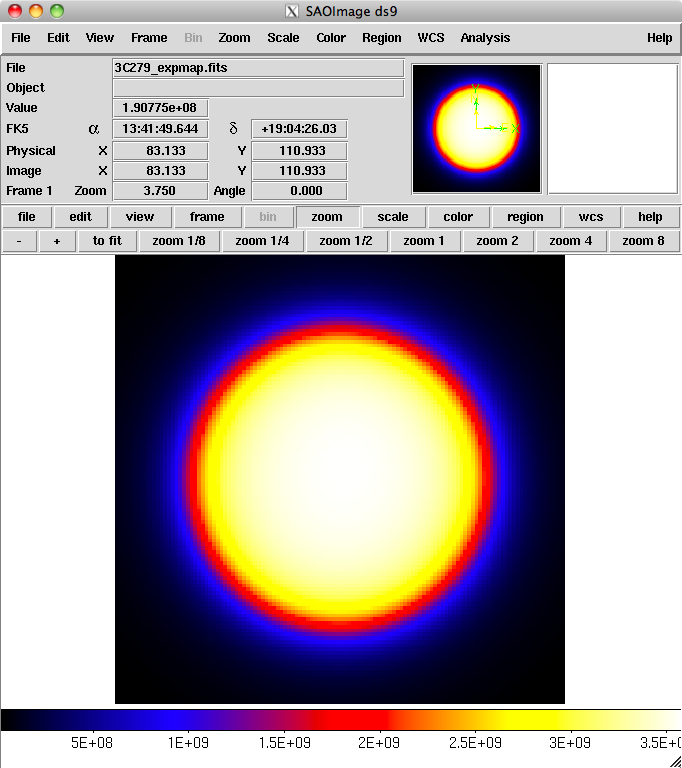
4. Download the latest background models
When you use the current Galactic diffuse emission model (gll_iem_v05_rev1.fit) in a likelihood analysis, you also want to use the corresponding model for the extragalactic isotropic diffuse emission, which includes the residual cosmic-ray background. The recommended isotropic model is iso_source05_rev1.txt.
Note: The Galactic diffuse model gll_iem_v05_rev1.fit is updated and had an additional components compared to the gll_iem_v05.fits Galactic model. The isotropic compenent is unchanged but the names have been updated to the consistent with the current model. Also the diffuse column for the isotropic component still has the name iso_source_v05.
For this example, you should save it in the same directory with your data. The isotropic spectrum is valid only for the latest response functions and only for data sets with front + back events combined. All of the most up-to-date background models along with a description of the models are available here.
5. Create a source model XML file.
The gtlike tool reads the source model from an XML file. The model file contains your best guess at the locations and spectral forms for the sources in your data. A source model can be created using the model editor tool, by using the user contributed tool make2FGLxml.py, or by editing the file directly within a text editor.
Given the dearth of bright sources in the extraction region we have selected, our source model file will be fairly simple, comprising only the Galactic and Extragalactic diffuse emission, and point sources to represent the blazars 3C 279 and 3C 273. Here is an example XML file:
<?xml version="1.0" ?>
<source_library title="source library">
<!-- Point Sources -->
<source name="3C 273" type="PointSource">
<spectrum type="PowerLaw">
<parameter free="1" max="1000.0" min="0.001" name="Prefactor" scale="1e-09" value="10"/>
<parameter free="1" max="-1.0" min="-5.0" name="Index" scale="1.0" value="-2.1"/>
<parameter free="0" max="2000.0" min="30.0" name="Scale" scale="1.0" value="100.0"/>
</spectrum>
<spatialModel type="SkyDirFunction">
<parameter free="0" max="360" min="-360" name="RA" scale="1.0" value="187.25"/>
<parameter free="0" max="90" min="-90" name="DEC" scale="1.0" value="2.17"/>
</spatialModel>
</source>
<source name="3C 279" type="PointSource">
<spectrum type="PowerLaw">
<parameter free="1" max="1000.0" min="0.001" name="Prefactor" scale="1e-09" value="10"/>
<parameter free="1" max="-1.0" min="-5.0" name="Index" scale="1.0" value="-2"/>
<parameter free="0" max="2000.0" min="30.0" name="Scale" scale="1.0" value="100.0"/>
</spectrum>
<spatialModel type="SkyDirFunction">
<parameter free="0" max="360" min="-360" name="RA" scale="1.0" value="193.98"/>
<parameter free="0" max="90" min="-90" name="DEC" scale="1.0" value="-5.82"/>
</spatialModel>
</source>
<!-- Diffuse Sources -->
<source name="gll_iem_v05_rev1" type="DiffuseSource">
<spectrum type="PowerLaw">
<parameter free="1" max="10" min="0" name="Prefactor" scale="1" value="1"/>
<parameter free="0" max="1" min="-1" name="Index" scale="1.0" value="0"/>
<parameter free="0" max="2e2" min="5e1" name="Scale" scale="1.0" value="1e2"/>
</spectrum>
<spatialModel file="./gll_iem_v05_rev1.fit" type="MapCubeFunction">
<parameter free="0" max="1e3" min="1e-3" name="Normalization" scale="1.0" value="1.0"/>
</spatialModel>
</source>
<source name="iso_source_v05" type="DiffuseSource">
<spectrum file="./iso_source_v05_rev1.txt" type="FileFunction">
<parameter free="1" max="10" min="1e-2" name="Normalization" scale="1" value="1"/>
</spectrum>
<spatialModel type="ConstantValue">
<parameter free="0" max="10.0" min="0.0" name="Value" scale="1.0" value="1.0"/>
</spatialModel>
</source>
</source_library>The XML file used for this example is 3C279_input_model.xml. For more details on the available XML models, see:
Descriptions of available Spectral and Spatial Models
Examples of XML Model Definitions for Likelihood
XML for Extended Sources
If your region includes any extended sources, you will need to add one or more extended sources to your XML model. Download the extended source templates from the LAT Catalog page (look for `Extended Source template archive'). Extract the archive in the directory of your choice and note the path to the template files, which have names like `W44.fits' and `VelaX.fits'. You will need to modify your XML model so that the path to the template file is correct. In addition, you will need to add the modifier map_based_integral=`true' to the <spatialModel> atttribute. This modifier tells gtlike to use the proper integration method when evaluating the extended source. (This modifier should set to false for Binned Likelihood.) You will also need to run gtdiffrsp in the next step to ensure that the diffuse response is properly calculated for that extended source.
Here is an example of the proper format for an extended source XML entry for Unbinned Likelihood analysis:
<?xml version="1.0" ?>
<source_library title="source library" xmlns="http://fermi.gsfc.nasa.gov/source_library">
<source name="3C 273" type="PointSource">
<spectrum type="PowerLaw">
<parameter free="1" max="1000.0" min="0.001" name="Prefactor" scale="1e-09" value="10"/>
<parameter free="1" max="-1.0" min="-5.0" name="Index" scale="1.0" value="-2.1"/>
<parameter free="0" max="2000.0" min="30.0" name="Scale" scale="1.0" value="100.0"/>
</spectrum>
<spatialModel type="SkyDirFunction">
<parameter free="0" max="360" min="-360" name="RA" scale="1.0" value="187.25"/>
<parameter free="0" max="90" min="-90" name="DEC" scale="1.0" value="2.17"/>
</spatialModel>
</source>
<source name="3C 279" type="PointSource">
<spectrum type="PowerLaw">
<parameter free="1" max="1000.0" min="0.001" name="Prefactor" scale="1e-09" value="10"/>
<parameter free="1" max="-1.0" min="-5.0" name="Index" scale="1.0" value="-2"/>
<parameter free="0" max="2000.0" min="30.0" name="Scale" scale="1.0" value="100.0"/>
</spectrum>
<spatialModel type="SkyDirFunction">
<parameter free="0" max="360" min="-360" name="RA" scale="1.0" value="193.98"/>
<parameter free="0" max="90" min="-90" name="DEC" scale="1.0" value="-5.82"/>
</spatialModel>
</source>
<!-- Diffuse Sources -->
<source name="gll_iem_v05_rev1" type="DiffuseSource">
<spectrum type="PowerLaw">
<parameter free="1" max="10" min="0" name="Prefactor" scale="1" value="1"/>
<parameter free="0" max="1" min="-1" name="Index" scale="1.0" value="0"/>
<parameter free="0" max="2e2" min="5e1" name="Scale" scale="1.0" value="1e2"/>
</spectrum>
<spatialModel file="./gll_iem_v05_rev1.fit" type="MapCubeFunction">
<parameter free="0" max="1e3" min="1e-3" name="Normalization" scale="1.0" value="1.0"/>
</spatialModel>
</source>
<source name="iso_source_v05" type="DiffuseSource">
<spectrum file="./iso_source_v05_rev1.txt" type="FileFunction">
<parameter free="1" max="10" min="1e-2" name="Normalization" scale="1" value="1"/>
</spectrum>
<spatialModel type="ConstantValue">
<parameter free="0" max="10.0" min="0.0" name="Value" scale="1.0" value="1.0"/>
</spatialModel>
</source>
</source_library>6. Compute the diffuse source responses.
Skip this step if you are using one of the recommended IRFs (CALDB) and the recommended diffuse background models, and your XML model does not include an extended source. The diffuse response for the standard analysis has been precomputed and is included in the event data file. You can find the current names of the diffuse response column using the FTOOLS with
fkeyprint <event file name>.fits DISRSP
e.g.
fkeyprint L14043015051399489F7F38_PH00'[1]'.fits DIFRSP
# FILE: L14043015051399489F7F38_PH00.fits[1]
# KEYNAME: DIFRSP
# EXTENSION: 1
DIFRSP0 = 'p7rep_source_v15__gll_iem_v05_rev1' / Diffuse response label for comp
DIFRSP1 = 'p7rep_source_v15__iso_source_v05' / Diffuse response label for compon
DIFRSP2 = 'p7rep_clean_v15__gll_iem_v05_rev1' / Diffuse response label for compo
DIFRSP3 = 'p7rep_clean_v15__iso_clean_v05' / Diffuse response label for componen
DIFRSP4 = 'NONE ' / Diffuse response label for component 4where the first part tells you the current IRF name and, after the double underscore, the name of the diffuse column. However, if you are using a different IRF or diffuse model, or if your source model includes an extended source, you MUST compute a custom diffuse response prior to continuing the analysis.
The gtdiffrsp tool compares the name in the XML model to the event file data header to determine the presence of precalculated diffuse response values. This means that you need to use the provided diffuse files and match the naming scheme used for the diffuse components in the example above in order to take advantage of the precalculated diffuse response.
If these quantities are not precomputed using the gtdiffrsp tool, then gtlike will compute them at runtime. However, if multiple fits and/or sessions with the gtlike tool are anticipated, it is probably wise to precompute these quantities, as this step is very computationally intensive often taking ~hours to complete. The source model XML file must contain all of the diffuse sources to be fit. The gtdiffrsp tool will add columns to the event data file for each diffuse source. Here we have copied the events file to a new file (3C279_events_gti_altdiff.fits) that will be used to calculate the alternate/non-standard diffuse response using an older IRF from P6V11.
prompt> gtdiffrsp
Event data file[] 3C279_events_gti_galtdiff.fits
Spacecraft data file[] spacecraft.fits
Source model file[] 3C279_input_model.xml
Response functions to use[] P6_V11_DIFFUSE
Working on...
3C279_events_gti_altdiff.fits.....................!
prompt> This file would now be available for performing a likelihood analysis if you wished to use the P6_V11_DIFFUSE instrument response function (IRF). There are a number of IRFs distributed with the Fermi Science Tools. Some of these response functions, like P7REP_SOURCE_V15::FRONT, are designed to address only a subset of the events in the dataset, and thus would require you to make additional cuts before they could be used.
7. Run gtlike.
We are now ready to run the gtlike application. You may need the results of the likelihood fit to be output in XML model form (e.g. to use in generating a test statistic map). To obtain an XML model output in addition to the standard results files, use the sfile parameter on the command line (as shown below) to designate the output XML model filename.
prompt> gtlike refit=yes plot=yes sfile=3C279_output_model.xml
Statistic to use (BINNED|UNBINNED) [] UNBINNED
Spacecraft file[] spacecraft.fits
Event file[] 3C279_region_filtered_gti.fits
Unbinned exposure map[] 3C279_expmap.fits
Exposure hypercube file[] 3C279_ltcube.fits
Source model file[] 3C279_input_model.xml
Response functions to use[] CALDB
Optimizer (DRMNFB|NEWMINUIT|MINUIT|DRMNGB|LBFGS) [] NEWMINUITMost of the entries prompted for are fairly obvious. In addition to the various XML and FITS files, the user is prompted for a choice of IRFs, the type of statistic to use, the optimizer, and some output file names.
The statistics available are:
UNBINNED This is a standard unbinned analysis, described in this tutorial, to be used for short timescale or low source count data. If this option is chosen then parameters for the spacecraft file, event file, and exposure file must be given.
BINNED This analysis is used for long timescale or high-density data (such as in the Galactic plane) which can cause memory errors in the unbinned analysis. See explanation in: Binned Likelihood Tutorial
There are five optimizers from which to choose: DRMNGB, DRMNFB, NEWMINUIT, MINUIT and LBFGS. Generally speaking, the faster way to find the parameter estimates is to use DRMNGB (or DRMNFB) to find initial values and then use MINUIT (or NEWMINUIT) to find more accurate results. If you have trouble achieving convergence at first, you can loosen your tolerance by setting the hidden parameter ftol on the command line. (The default value for ftol is 0.01.)
The application proceeds by reading in the spacecraft and event data, and if necessary, computing event responses for each diffuse source.
Here is the output from our fit:
Minuit did successfully converge.
# of function calls: 118
(MUCH OUTPUT OMITTED.)
Computing TS values for each source (4 total)
....!
Photon fluxes are computed for the energy range 100 to 100000 MeV
3C 273:
Prefactor: 9.73264 +/- 0.319754
Index: -2.62278 +/- 0.0240336
Scale: 100
Npred: 4726.06
ROI distance: 10.4409
TS value: 5623.03
Flux: 6.00384e-07 +/- 1.40167e-08 photons/cm^2/s
3C 279:
Prefactor: 8.26767 +/- 0.241538
Index: -2.22069 +/- 0.0161135
Scale: 100
Npred: 5962.5
ROI distance: 0
TS value: 10957.6
Flux: 6.77554e-07 +/- 1.36379e-08 photons/cm^2/s
gll_iem_v05_rev1:
Prefactor: 1.29472 +/- 0.0133664
Index: 0
Scale: 100
Npred: 64749.6
Flux: 0.000640583 +/- 6.61329e-06 photons/cm^2/s
iso_source_v05:
Normalization: 1.1256 +/- 0.015276
Npred: 50279.7
Flux: 0.00020212 +/- 2.74098e-06 photons/cm^2/s
WARNING: Fit may be bad in range [281.838, 562.341] (MeV)
Total number of observed counts: 125718
Total number of model events: 125718
-log(Likelihood): 1374139.948
Writing fitted model to 3C279_output_model.xmlSince we selected `plot=yes' in the command line, a plot of the fitted data appears.
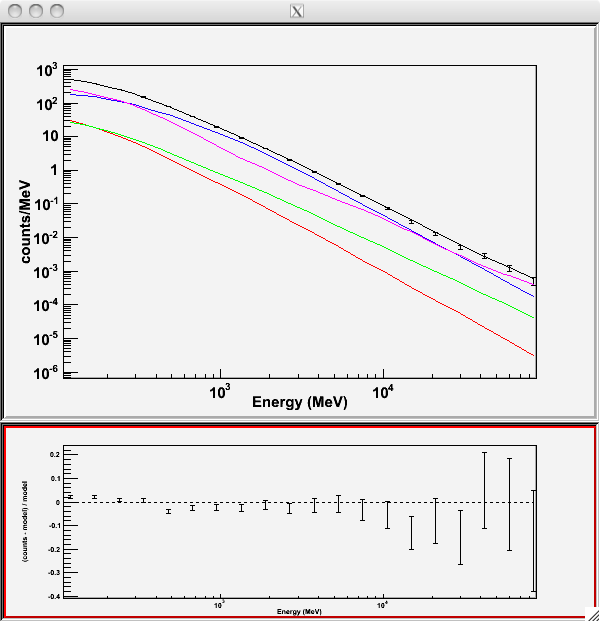
In the first plot the counts/MeV vs MeV are plotted. The points are the data, and the lines are the models. Error bars on the points represent sqrt(Nobs) in that band, where Nobs is the observed number of counts. The black line is the sum of the models for all sources. The colored lines follow the sources as follows:
Black - summed model
Red - first source in results.dat file (see below)
Green - second source
Blue - third source
Magenta - fourth source
Cyan - the fifth source
In our case the 3C273 is the one in red, 3C279 is the one in green, the extragalactic background is blue and the galactic background is magenta. If you have more sources the colors are reused in the same order.
The second plot gives the residuals between your model and the data. Error bars here represent (sqrt(Nopbs))/Npred, where Npred is the predicted number of counts in each band based on the fitted model.
To assess the quality of the fit, look first for the words at the top of the output `<Optimizer> did successfully converge.'. Successful convergence is a minimum requirement for a good fit. Next, look at the energy ranges that are generating warnings of bad fits. If any of these ranges affect your source of interest, you may need to revise the source model and refit. You can also look at the residuals on the plot (bottom panel). If the residuals indicate a poor fit overall, you should consider changing your model file, perhaps by using a different source model definition, and refit the data.
If the fits and spectral shapes are good, but could be improved, you may wish to simply update your model file to hold some of the spectral parameters fixed. For example, by fixing the spectral model for 3C 273, you may get a better quality fit for 3C 279. Close the plot and you will be asked if you wish to refit the data.
Refit? [y]n
Elapsed CPU time: 111.819315
prompt>Here, hitting `return' will instruct the application to fit again. We are happy with the result, so we type `n' and end the fit.
Results
When it completes, gtlike generates two standard output files: results.dat with the results of the fit, and counts_spectra.fits with predicted counts for each source for each energy bin. The points in the plot above are the data points from the counts_spectra.fits file, while the colored lines follow the source model information in the results.dat file. If you re-run the tool in the same directory, these files will be overwritten by default. Use the clobber=no option on the command line to keep from overwriting the output files.
Interestingly, the fit details and the value for the -log(likelihood) are not recorded in the automatic output files. You should consider logging the output to a text file for your records.
In this example, we used the `sfile' parameter to request that the model results be written to an output XML file, 3C279_output_model.xml. This file contains the source model results that were written to results.dat at the completion of the fit.
Note: If you have specified an output XML model file and you wish to modify your model while waiting at the `Refit? [y]' prompt, you will need to copy the results of the output model file to your input model before making those modifications.
Errors reported with each value in the results.dat file are 1σ estimates (based on the inverse-Hessian at the optimum of the log-likelihood surface).
Other Useful Hidden Parameters
If you are scripting and wish to generate multiple output files without overwriting, the 'results' and `specfile' parameters allow you to specify output filenames for the results.dat and counts_spectra.fits files respectively.
If you do not specify a source model output file with the `sfile' parameter, then the input model file will be overwritten with the latest fit. This is convenient as it allows the user to edit that file while the application is waiting at the `Refit? [y]' prompt so that parameters can be adjusted and set free or fixed. This would be similar to the use of the `newpar', `freeze', and `thaw' commands in XSPEC.
8. Make test-statistic maps.
Ultimately, one would like to find sources near the detection limit of the instrument. To do this, you model the strongest, most obvious sources (with some theoretical prejudice as to the true source positions, e.g., assuming that most variable high Galactic latitude sources are blazars which can be localized by radio, optical, or X-ray observations), and then create `Test-statistic maps' to search for unmodeled point sources. These TS maps are created by moving a putative point source through a grid of locations on the sky and maximizing -log(likelihood) at each grid point, with the other, stronger, and presumably well-identified sources included in each fit. New, fainter sources are then identified at local maxima of the TS map.
The gttsmap tool can be run with or without an input source model. However, for a useful visualization of the results of the fit, it is recommended you use the output model file from gtlike. The file must be edited so all parameters are fixed (by setting the `free' attribute to 0 for each parameter). Otherwise gttsmap will attempt a refit of the entire model at every point on the grid. To see the field with the fitted sources removed (i.e. a residuals map), fix all point source parameters before running the TS map. To see the field with the fitted sources included, edit the model to remove all but the diffuse components.
For residuals map: 3C279_output_model_resid.xml
For sources TS map: 3C279_output_model_diffonly.xml
In both cases, leave the Galactic diffuse prefactor (called `Value' in the model file) and the isotropic diffuse normalization parameters free during the fit.
Running gttsmap is extremely time-consuming, as the tool is performing a likelihood fit for all events at every pixel position. One way to reduce the time required for this step is to use very coarse binning and/or a very small region. In the following example, we run a TS map for the central 20x20 degree region of our data file, with .25 degreebins. This results in 6400 maximum likelihood calculations. The run time for each of the maps discussed below was approximately four days.
Here is an example of how to run the gttsmap tool to look for additional sources:
prompt> gttsmap
Event data file[] 3C279_region_filtered_gti.fits
Spacecraft data file[] spacecraft.fits
Exposure map file[none] 3C279_expmap.fits
Exposure hypercube file[] 3C279_ltcube.fits
Source model file[] 3C279_output_model_resid.xml
TS map file name[] 3C279_tsmap_resid.fits
Response functions to use[] CALDB
Optimizer (DRMNFB|NEWMINUIT|MINUIT|DRMNGB|LBFGS) [] NEWMINUIT
Fit tolerance[] 1e-5
Number of X axis pixels[] 80
Number of Y axis pixels[] 80
Image scale (in degrees/pixel)[] 0.25
Coordinate system (CEL|GAL) [] CEL
X-coordinate of image center in degrees (RA or l)[] 193.98
Y-coordinate of image center in degrees (Dec or b)[] -5.82
Projection method (AIT|ARC|CAR|GLS|MER|NCP|SIN|STG|TAN) [] AIT
.....................!The output from the fit is below. Because generating TS maps takes a long time, you may wish to download the residuals and source files attached here. In the left panel, only diffuse sources were included in the analysis. The right panel shows the same field, but with the point sources (3C 279 and 3C 273) included in the model, and thus not included in the output image. This gives a pseudo-residuals map. The bright source clearly seen in the lower part of this image is a recently identified millisecond pulsar. The location of the cursor in the image indicates that this source had a TS of 1608 for the first six months of the mission.
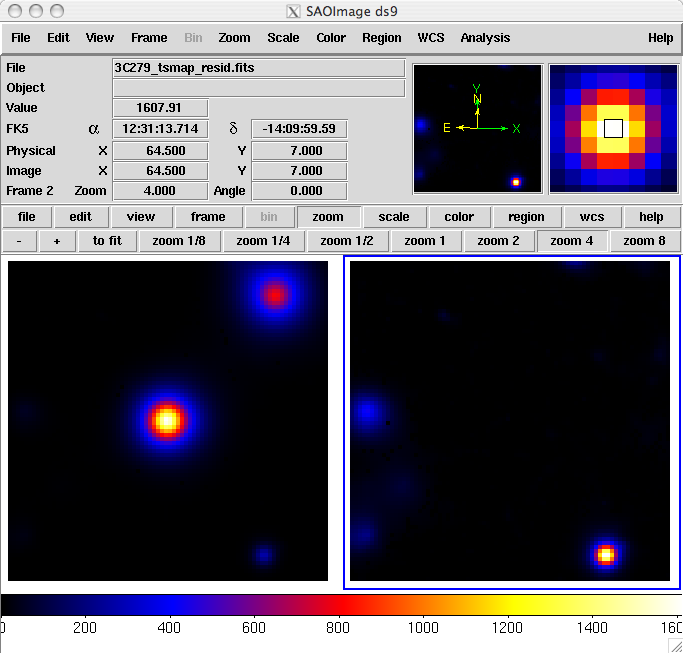
In this example, the data set was small enough that an unbinned likelihood analysis is possible. With longer data sets, however, you may run into memory allocation errors, at which time it is necessary to move to binned analysis for your region. A typical memory allocation error looks like:
gtlike(10869) malloc: *** mmap(size=2097152) failed (error code=12)*** error: can't allocate region*** set a breakpoint in malloc_error_break to debug
If you encounter this error, then Binned Likelihood Analysis is for you!
Last updated by: D. Davis 05/07/2014



